Developing SmartSense: A Robotic Platform for High Throughput Phenotyping and Genome Wide Association Analysis
The overarching vision of this proposal is to usher in Big Agriculture Data techniques by developing a versatile sensing platform, termed SmartSense, that can be mounted on robots to collect vast scales of data from fields (Figure 1). The specific objectives of this project are two-fold. (1) The agriculture goal is to improve soybean productivity through genotyping and high-throughput phenotyping. (2) The technology goal is to develop SmartSense and demonstrate its utility by collecting large scales of data through operations with unmanned aerial vehicles (UAVs) and ground vehicles (UGVs). Our research team includes faculty from CALS and COE with complementary expertise in genomics (Li), soybean breeding (Zhang), and robotics (Tokekar) covering all technical aspects of the proposed efforts.
Soybean is a major crop grown in more than 80 million acres in the US and is widely used in multiple industries including food production, bio-diesel production and animal feeding. Improvement of soybean yield is lagging behind other major crops such as maize and wheat due to the limited yield potential. A promising direction to improve soybean productivity is to breed soybean varieties with better plant architecture which can lead to improved harvest efficiency, reduced cost for weed management, and higher yield potential.
We will develop innovative robotic technology which can collect phenotypic data from the field. Computer vision and machine learning algorithms will be developed to recognize flowers, pods, leaves and weeds from images collected. Genomic data and statistical analysis will be performed to identify key genetic variations that associated with these desirable traits.
The agriculture goal of this proposal is to identify genetic markers that are associated with plant architecture traits such as leaf angle, uniform flowering time, increased pod to flower ratio, faster canopy closure and higher pod location. Identification of these markers will enable breeding of new soybean varieties with improved productivity. For example, optimal leaf angle can improve photosynthetic efficiency; Uniform flowering time and increased pod to flower ratio can increase yield potential and decrease management cost; Faster canopy closure provides competitive advantage of crops over weeds and can reduce herbicide usage and limit its environmental impact; Higher pod location improves harvest efficiency for mechanical harvesters.
Traditionally, breeding for plant architecture has been limited by the cost of labor for performing field phenotyping. Only simple traits such as seed weights and protein/oil contents were used as target traits because these traits are easy to measure after harvest. In recent years, reduced cost ingenomic technology has provided genotypes of tens of thousands of soybean varieties[1]. These genotyped soybean varieties provide a rich genetic pool for breeders to select genomic markers that associated better plant architecture traits. At the same time, innovative robotic technologies allow for rapid and cost-effective collection of plant phenotypic data in field conditions[2]. Improved computer vision algorithms and reduced cost for data storage have also enabled collection and analysis of large scale field phenotyping data during the growing season. This project comes at an opportune time to synergistically exploit the advances in both fields.
In this proposal, we will improve our existing aerial imaging platform with multi-spectral sensorssources (RGB, Near-IR, thermal, LiDAR) to collect leaf coverage data, plant nutrient status and weed pressure in the field. We also plan to develop ground based, field phenotyping robot that can collect data for soybeans by traversing between the crop rows, because soybean is a broad leaf crop, aerial images cannot detect flower and pod locations. One of the outputs from the data collected by the ground robot will be a 3D reconstruction of the structure of soybean plants. Using advance computer vision and machine learning algorithms, we will identify flower and pod locations, count number of flowers and pods, calculate canopy growth curve and identify weeds from both aerial and ground-based images. Some of these results will be used as quantitative trait to perform GWAS study and to identify genomic markers that associated with these traits. These genomic markers will then be used to selectively breed new soybean varieties.
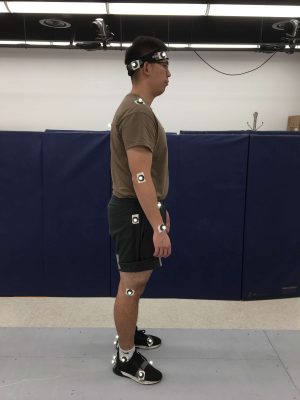
We are funded by Virginia Tech SmartFarm initiative to construct SmartSense, a light-weight, multi-spectral imaging platform. The ICAT project, if funded, will enable integration of the SmartSense platform on aerial and ground robots, development of machine learning and computer vision techniques for producing outputs such as detailed 3D reconstruction of plant architecture and health monitoring, and leveraging the platform for improving soybean productivity. This sensor platform is designed to following a recently published protocol [2] using updated hardware (Figure 1). It will include a laser scanner (LiDAR) for detailed reconstruction of plant morphology and sensors for measuring Normalized Difference Vegetation Index (NDVI). Data will be logged using a credit-card sized computing platform (Nvidia, Jetson TX1) with powerful GPU for onboard real-time processing. SmartSense will be small enough to be mounted on an unmanned ground vehicle (UGV, Lynxmotion A2WD) and an unmanned aerial vehicle (UAV, DJI S900) present in Tokekar’s lab.
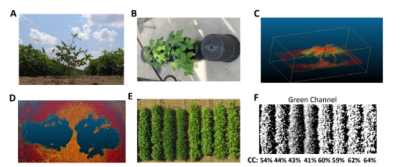
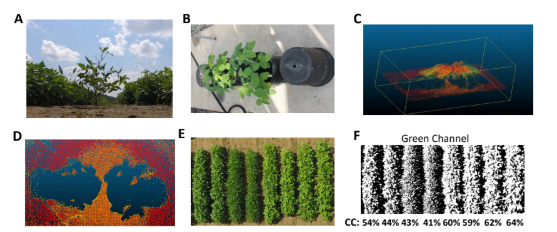
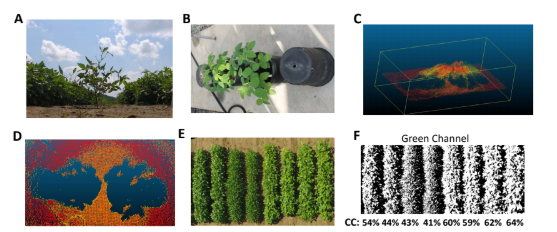
For phenotyping of pod and flower locations, the UGV will be used to collected RGB images, point cloud and NDVI images from the camera mounted ~2 inches above ground (Figure 2 A). The RGB images will be used for manual labeling of pod and flower locations and will be used as positive training samples. We will apply multiple machine learning tools including support vector machine [3], random forest [4] and deep learning neural networks to recognize the objects from these images [5,6]. Based on the image analysis results, we will calculate the number of pods and flowers at different distance from the ground. Similar machine learning strategy will be used to identify the locations of weeds from aerial images collected by SmartSense mounted on the UAV.
For phenotyping of canopy closure, we will apply fast aerial phenotyping platform using a UAV carrying the SmartSense sensor. Images will be collected using pre-programed route that is designed based on the geometry of the testing farm. We will collect three types of data for canopy coverage: crown projection area using LiDAR[7], see Figure 2 B-D for example; canopy coverage as percentage of pixels classified as canopy[8], see Figure 2 E-F, and Normalized Difference Vegetation Index (NDVI)[9]. These indices for canopy coverage will be calibrated using ground-based images. Growth curves will be fitted to the canopy coverage data. Ground based images for pod locations will be taken before harvesting. Quantitative traits collected from the images will be used in GWAS analysis to identify genomic markers that significantly associated with these traits. We will apply our published protocol in GWAS analysis for soybeans[10]. In brief, quantitative traits such as leaf angle and pod height will be collected for the GWAS population. Mixed linear model will be used for association study that controls for confounding factors such as hidden population structure and kinship relation in this population. Candidate SNPs will be selected based on p value and LOD values. Candidate genes will be identified based on gene function annotation that are related to traits of interest.
The goals of this project are two-fold: (1) To develop a flexible sensing platform that can collect large scales of phenotypic data using unmanned aerial and ground robots; and (2) apply phenotypic data and use GWAS to identify genomic variations and markers that are associated with traits of interest to improve soybean production.
In addition to the outcomes described in the previous section, the main deliverables of the project will be computer vision and machine learning algorithms for translating raw agriculture outputs to actionable data products, navigation algorithms for robots to collect phenotypic data, and pipelines to identify genetic markers using phenomic data collected by robots.
ICAT 2018-2019 Major Sead Grant