Visual Language of Chromatin Architecture

It is now well recognized that three-dimensional (3D) organization of the genome (chromatin) plays an important role in key cellular processes such as DNA replication, repair, transcription, and epigenetic inheritance, i.e., inheritance that is not encoded by the DNA sequence. Abnormalities in the 3D genome architecture have been associated with a number of pathologies including various cancers. Yet the details of the 3D architecture are only beginning to emerge: what is clear is that the relevant structures are exceedingly complex, possibly including various fractal forms – these are not easy to comprehend and connect to biological function. The problem is exacerbated by the lack of powerful, information-reach domain-specific visualization tools, proved to be so useful in many other fields. The 3D objects relevant to the chromatin field are so inherently complex that “standard” visualization tools and structure representation concepts borrowed from the field of structural biology (proteins, DNA) are likely to be inadequate to support the new field. New, frontier tools and concepts, and likely a new language to represent these structures are needed, aided by powerful computation. At the same time, this new field of science offers unique opportunity to explore and develop novel art forms, where art meets science and computation. In all these respects, ICAT offers a unique opportunity for this pilot project.
The broad goals of this proposal are two-fold: (1) Develop a set of tools and artistic interpretations that will aid conceptual understanding and, eventually, detailed reasoning, of the hierarchy of DNA packing inside the nucleus. This very tight packing – several feet of the DNA must fit inside the nucleus of only a few micrometers in size (Figure 1, left) – is critical for the function of the genome. The packing is likely to involve very complex geometries, including fractal (Figure 1, middle). Artistic vision has been known to help conceptual understanding (Figure 1, right). (2) Conversely, the beauty of fundamental laws of Nature has inspired great artworks from the ancient Greeks to Da Vinci to Dali. The mysterious yet fundamental geometrical objects relevant to chromatin biology will inspire new art forms that we will develop in this project.


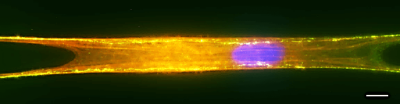
We will develop computational models of chromosome organization in nuclei for polytene chromosomes, focusing on ways to visualize the most relevant information. We will validate the models against the corresponding experimental data at the appropriate resolution. To facilitate the tracing of individual polytene chromosomal arms, to visualize structure of bands and to identify chromosoma territories, we will use labeled DNA, proteins and chromosome painting probes as shown in Figure 2.

We will develop a visualization format/data structure prototype for dynamic access to the multiple scales of nuclear architecture and the corresponding layers of annotation. The idea is to map the chromosome structural hierarchy, e.g., gene < locus < chromosome < territory, onto the macromolecular structural hierarchy already supported by many well-designed software packages. For example, a single gene can be associated with an atom, which can be of several types (colors), while the whole chromosome may be associated with a protein chain and its attributes (Figure 3). As our visualization platform, we will use one of the most popular and versatile free software packages in structural biology – Visual Molecular Dynamics (VMD). The package has nearly 200,000 users and is designed to visualize and manipulate a wealth of hierarchical information found in macromolecules. This platform will provide excellent solutions to the similar problem of displaying and manipulating macromolecular structures on multiple scales (e.g., atom < amino-acid < protein subunit < protein complex). In this endeavor we will be receiving consultation from the VMD development team (UIUC) and co-PI Onufriev’s colleagues at the CS department (VT) directly associated with the CUBE. A talented CS undergraduate student (Raju Nadimpalli ) is already working with our team on chromatin visualization; this funding will help us retain him for the summer and beyond.
The studio work will employ a nimble production approach utilizing (but not limited to) laser-cutting techniques, photography, 3D printing, gilding, sculpting and woodworking. Techniques, processes and aesthetics will be determined by the conceptual interpretation and scientific understanding of DNA packing, therefor allowing the work to come to fruition with as little predetermination as possible. The combined use of technology in production is often relinquished to processing and algorithmic computation. Our intention here is to incorporate the biological from conception to production process. Visual-result processing and algorithms are to be replaced by artistic sensibility in as much capacity as possible.